Developer documentation¶
Scikit-multilearn development team is an open international community that welcomes contributions and new developers. This document is for you if you want to implement a new:
- classifier
- relationship graph builder
- label space clusterer
Before we can go into development details, we need to discuss how to setup a comfortable development environment and what is the best way to contribute.
Working with the repository¶
Scikit-learn is developed on github using git for code version management. To get the current codebase you need to checkout the scikit-multilearn repository
git clone git@github.com:scikit-multilearn/scikit-multilearn.git
To make a contribution to the repository your should fork the
repository, clone your fork, and start development based on the
master branch. Once you’re done, push your commits to your
repository and submit a pull request for review.
The review usually includes: - making sure that your code works, i.e. it has enough unit tests and tests pass - reading your code’s documentation, it should follow the numpydoc standard - checking whether your code works properly on sparse matrix input - your class should not store more data in memory than neccessary
Once your contributions adhere to reviewer comments, your code will be included in the next release.
Development Docker image¶
To ease development and testing we provide a docker image containing all libraries needed to test all of scikit-multilearn codebase. It is an ubuntu based docker image with libraries that are very costly to compile such as python-graphtool. This docker image can be easily integrated with your PyCharm environment.
To pull the scikit-multilearn docker image just use:
$ docker pull niedakh/scikit-multilearn-dev:latest
After cloning the scikit-multilearn repository, run the following command:
This docker contains two python environments set for scikit-multilearn:
2.7 and 3.x, to use the first one run python2 and pip2, the
second is available via python3 and pip3.
You can pull the latest version from Docker hub using:
$ docker pull niedakh/scikit-multilearn-dev:latest
You can start it via:
$ docker run -e "MEKA_CLASSPATH=/opt/meka/lib" -v "YOUR_CLONE_DIR:/home/python-dev/repo" --name scikit_multilearn_dev_test_docker -p 8888:8888 -d niedakh/scikit-multilearn-dev:latest
To run the tests under the python 2.7 environment use:
$ docker exec -it scikit_multilearn_dev_test_docker python3 -m pytest /home/python-dev/repo
or for python 3.x use:
$ docker exec -it scikit_multilearn_dev_test_docker python2 -m pytest /home/python-dev/repo
To play around just login with:
$ docker exec -it scikit_multilearn_dev_test_docker bash
To start jupyter notebook run:
$ docker exec -it scikit_multilearn_dev_test_docker bash -c "cd /home/python-dev/repo && jupyter notebook"
Building documentation¶
In order to build HTML documentation just run:
$ docker exec -it scikit_multilearn_dev_test_docker bash -c "cd /home/python-dev/repo/docs && make html"
Development¶
One of the most comfortable ways to work on the library is to use
Pycharm and its support for
docker-contained
interpreters,
just configure access to the docker server, set it up in Pycharm, use
niedakh/scikit-multilearn-dev:latest as the image name and set up
relevant path mappings, voila - you can now use this environment for
development, debugging and running tests within the IDE.
Writing code¶
At the very list you should make sure that your code:
- works on Python 2 and Python 3 on Windows 10/Linux/OSX using travis/appveyor
- PEP8 coding guidelines
- follows scikit-learn interfaces if relevant interfaces exist
- is documented in the numpydocs fashion, especially that all public API is documented, including attributes and an example use case, see existing code for inspiration
- has tests written, you can find relevant tests in
skmultilearn.cluster.testsandskmultilearn.problem_transform.tests.
Writing a label space clusterer¶
One of the approaches to multi-label classification is to cluster the label space into subspaces and perform classification in smaller subproblems to reduce the risk of under/overfitting.
In order to create your own label space clusterer you need to inherit
:class:LabelSpaceClustererBase and implement the
fit_predict(X, y) class method. Expect X and y to be sparse
matrices, you and also use
:func:skmultilearn.utils.get_matrix_in_format to convert to a
desired matrix format. fit_predict(X, y) should return an array-like
(preferably ndarray or at least a list) of n_clusters
subarrays which contain lists of labels present in a given cluster. An
example of a correct partition of five labels is:
np.array([[0,1], [2,3,4]]) and of overlapping clusters:
np.array([[0,1,2], [2,3,4]]).
Example Clusterer¶
Let us look at a toy example, where a clusterer divides the label space based on how a given label’s ordinal divides modulo a given number of clusters.
In [1]:
from skmultilearn.dataset import load_dataset
In [2]:
X_train, y_train, _, _ = load_dataset('emotions', 'train')
X_test, y_test, _, _ = load_dataset('emotions', 'test')
emotions:train - exists, not redownloading
emotions:test - exists, not redownloading
In [81]:
import numpy as np
from skmultilearn.ensemble import LabelSpacePartitioningClassifier
from skmultilearn.cluster.base import LabelSpaceClustererBase
class ModuloClusterer(LabelSpaceClustererBase):
"""Initializes the clusterer
Parameters
----------
n_clusters: int
number of clusters to partition into
Returns
--------
array-like of array-like, (n_clusters,)
list of lists label indexes, each sublist represents labels
that are in that community
"""
def __init__(self, n_clusters = None):
super(ModuloClusterer, self).__init__()
self.n_clusters = n_clusters
def fit_predict(self, X, y):
n_labels = y.shape[1]
partition_list = [[] for _ in range(self.n_clusters)]
for label in range(n_labels):
partition_list[label % self.n_clusters].append(label)
return np.array(partition_list)
In [13]:
clusterer = ModuloClusterer(n_clusters=3)
clusterer.fit_predict(X_train, y_train)
Out[13]:
array([[0, 3],
[1, 4],
[2, 5]])
Using the example Clusterer¶
Such a clusterer can then be used with an ensemble classifier such as
the LabelSpacePartitioningClassifier.
In [14]:
from skmultilearn.ensemble import LabelSpacePartitioningClassifier
from skmultilearn.problem_transform import LabelPowerset
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
In [15]:
clf = LabelSpacePartitioningClassifier(
classifier = LabelPowerset(classifier=GaussianNB()),
clusterer = clusterer
)
clf
Out[15]:
LabelSpacePartitioningClassifier(classifier=LabelPowerset(classifier=GaussianNB(priors=None), require_dense=[True, True]),
clusterer=ModuloClusterer(n_clusters=3),
require_dense=[False, False])
In [16]:
clf.fit(X_train, y_train)
prediction = clf.predict(X_test)
accuracy_score(y_test, prediction)
Out[16]:
0.23762376237623761
Writing a Graph Builder¶
Scikit-multilearn implements clusterers that are capable of infering label space clusters (in network science the word communities is used more often) from a graph/network depicting label relationships. These clusterers are further described in Label relations chapter of the user guide.
To implement your own graph builder you need to subclass
GraphBuilderBase and implement the transform function which
should return a weighted (or not) adjacency matrix in the form of a
dictionary, with keys (label1, label2) and values representing a
weight.
Example GraphBuilder¶
Let’s implement a simple graph builder which returns the correlations between labels.
In [58]:
from scipy import stats
from skmultilearn.cluster import GraphBuilderBase
from skmultilearn.utils import get_matrix_in_format
class LabelCorrelationGraphBuilder(GraphBuilderBase):
"""Builds a graph with label correlations on edge weights"""
def transform(self, y):
"""Generate weighted adjacency matrix from label matrix
This function generates a weighted label correlation
graph based on input binary label vectors
Parameters
----------
y : numpy.ndarray or scipy.sparse
dense or sparse binary matrix with shape
``(n_samples, n_labels)``
Returns
-------
dict
weight map with a tuple of ints as keys
and a float value ``{ (int, int) : float }``
"""
label_data = get_matrix_in_format(y, 'csc')
labels = range(label_data.shape[1])
self.is_weighted = True
edge_map = {}
for label_1 in labels:
for label_2 in range(0, label_1+1):
# calculate pearson R correlation coefficient for label pairs
# we only include the edges above diagonal as it is an undirected graph
pearson_r, _ = stats.pearsonr(label_data[:,label_2].todense(), label_data[:,label_1].todense())
edge_map[(label_2, label_1)] = pearson_r[0]
return edge_map
In [49]:
graph_builder = LabelCorrelationGraphBuilder()
In [50]:
graph_builder.transform(y_train)
Out[50]:
{(0, 0): 1.0,
(0, 1): 0.0054205072520802679,
(0, 2): -0.4730507042031965,
(0, 3): -0.35907118960632034,
(0, 4): -0.32287762681546733,
(0, 5): 0.24883125852376733,
(1, 1): 1.0,
(1, 2): 0.1393556218283642,
(1, 3): -0.25112700233108359,
(1, 4): -0.3343594619173676,
(1, 5): -0.36277277605002756,
(2, 2): 1.0,
(2, 3): 0.34204580629202336,
(2, 4): 0.23107157941324433,
(2, 5): -0.56137098197912705,
(3, 3): 1.0,
(3, 4): 0.48890609122000817,
(3, 5): -0.35949125643829821,
(4, 4): 1.0,
(4, 5): -0.28842101609587079,
(5, 5): 1.0}
This adjacency matrix can be then used by a Label Graph clusterer.
Using the example GraphBuilder¶
In [56]:
from skmultilearn.cluster import NetworkXLabelGraphClusterer
clusterer = NetworkXLabelGraphClusterer(graph_builder=graph_builder)
clusterer.fit_predict(X_train, y_train)
Out[56]:
array([[0, 5], [1], [2], [3, 4]], dtype=object)
The clusterer can be then used with the LabelSpacePartitioning classifier.
In [57]:
from skmultilearn.ensemble import LabelSpacePartitioningClassifier
from skmultilearn.problem_transform import LabelPowerset
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
clf = LabelSpacePartitioningClassifier(
classifier = LabelPowerset(classifier=GaussianNB()),
clusterer = clusterer
)
clf.fit(X_train, y_train)
prediction = clf.predict(X_test)
accuracy_score(y_test, prediction)
Out[57]:
0.13861386138613863
Writing a classifier¶
To implement a multi-label classifier you need to subclass a classifier base class. Currently, you can select of a few classifier base classes depending on which approach to multi-label classification you follow.
Scikit-multilearn inheritance tree for the classifier is shown on the figure below.
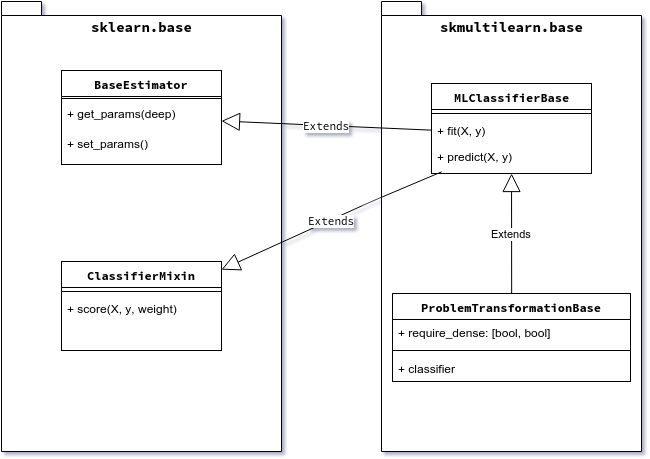
Classifier inheritance diagram
To implement a scikit-learn’s ecosystem compatible classifier, we need
to subclass two classes from sklearn.base: BaseEstimator and
ClassifierMixin. For that we provide
:class:skmultilearn.base.MLClassifierBase base class. We further
extend this class with properties specific to the problem transformation
approach in multi-label classification in
:class:skmultilearn.base.ProblemTransformationBase.
To implement a scikit-learn’s ecosystem compatible classifier, we need
to subclass two classes from sklearn.base: BaseEstimator and
ClassifierMixin. For that we provide
:class:skmultilearn.base.MLClassifierBase base class. We further
extend this class with properties specific to the problem transformation
approach in multi-label classification in
:class:skmultilearn.base.ProblemTransformationBase.
Scikit-learn base classses¶
The base estimator class from scikit is responsible for providing the ability of cloning classifiers, for example when multiple instances of the same classifier are needed for cross-validation performed using the CrossValidation class.
The class provides two functions responsible for that: get_params,
which fetches parameters from a classifier object and set_params,
which sets params of the target clone. The params should also be
acceptable by the constructor.
This is an interface with a non-important method that allows different
classes in scikit to detect that our classifier behaves as a classifier
(i.e. implements fit/predict etc.) and provides certain kind of
outputs.
MLClassifierBase¶
The base multi-label classifier in scikit-multilearn is
:class:skmultilearn.base.MLClassifierBase. It provides two abstract
methods: fit(X, y) to train the classifier and predict(X) to predict
labels for a set of samples. These functions are expected from every
classifier. It also provides a default implementation of
get_params/set_params that works for multi-label classifiers.
All you need to do in your classifier is:
- subclass
MLClassifierBaseor a derivative class - set
self.copyable_attrsin your class’s constructor to a list of fields (as strings), that should be cloned (usually it is equal to the list of constructor’s arguments) - implement the
fitmethod that trains your classifier - implement the
predictmethod that predicts results
One of the most important concepts in scikit-learn’s BaseEstimator,
is the concept of cloning. Scikit-learn provides a plethora of
experiment performing methods, among others, cross-validation, which
require the ability to clone a classifier. Scikit-multilearn’s base
multi-label class - MLClassifierBase - provides infrastructure for
automatic cloning support.
An example of this would be:
from skmultilearn.base import MLClassifierBase
class AssignKBestLabels(MLClassifierBase):
"""Assigns k most frequent labels
Parameters
----------
k : int
number of most frequent labels to assign
Example
-------
An example use case for AssignKBestLabels:
.. code-block:: python
from skmultilearn.<YOUR_CLASSIFIER_MODULE> import AssignKBestLabels
# initialize LabelPowerset multi-label classifier with a RandomForest
classifier = AssignKBestLabels(
k = 3
)
# train
classifier.fit(X_train, y_train)
# predict
predictions = classifier.predict(X_test)
"""
def __init__(self, k = None):
super(AssignKBestLabels, self).__init__()
self.k = k
self.copyable_attrs = ['k']
The fit(self, X, y) expects classifier training data:
Xshould be a sparse matrix of shape:(n_samples, n_features), although for compatibility reasons array of arrays and a dense matrix are supported.yshould be a sparse, binary indicator, matrix of shape:(n_samples, n_labels)with 1 in a positioni,jwheni-th sample is labelled with label no.j
It should return self after the classifier has been fitted to
training data. It is customary that fit should remember n_labels
in a way. In practice we store n_labels as self.label_count in
scikit-multilearn classifiers.
Let’s make our classifier trainable:
def fit(self, X, y):
"""Fits classifier to training data
Parameters
----------
X : `array_like`, :class:`numpy.matrix` or :mod:`scipy.sparse` matrix, shape=(n_samples, n_features)
input feature matrix
y : `array_like`, :class:`numpy.matrix` or :mod:`scipy.sparse` matrix of `{0, 1}`, shape=(n_samples, n_labels)
binary indicator matrix with label assignments
Returns
-------
self
fitted instance of self
"""
frequencies = (y_train.sum(axis=0)/float(y_train.sum().sum())).A.tolist()[0]
labels_sorted_by_frequency = sorted(range(y_train.shape[1]), key = lambda i: frequencies[i])
self.labels_to_assign = labels_sorted_by_frequency[:self.k]
return self
The predict(self, X) returns a prediction of labels for the samples
from X:
Xshould be a sparse matrix of shape:(n_samples, n_features), although for compatibility reasons array of arrays and a dense matrix are supported.
The returned value is similar to y in fit. It should be a sparse
binary indicator matrix of the shape (n_samples, n_labels).
In some cases, while scikit continues to progress towards a complete
switch to sparse matrices, it might be needed to convert the sparse
matrix to a dense matrix or even array-like of array-likes. Such
is the case for some scoring functions in scikit. This problem should go
away in the future versions of scikit.
The predict_proba(self, X) functions similarly but returns the
likelihood of the label being correctly assigned to samples from X.
Let’s add the prediction functionality to our classifier and see how it works:
In [99]:
from skmultilearn.base import MLClassifierBase
from scipy.sparse import lil_matrix
class AssignKBestLabels(MLClassifierBase):
"""Assigns k most frequent labels
Parameters
----------
k : int
number of most frequent labels to assign
Example
-------
An example use case for AssignKBestLabels:
.. code-block:: python
from skmultilearn.<YOUR_CLASSIFIER_MODULE> import AssignKBestLabels
# initialize LabelPowerset multi-label classifier with a RandomForest
classifier = AssignKBestLabels(
k = 3
)
# train
classifier.fit(X_train, y_train)
# predict
predictions = classifier.predict(X_test)
"""
def __init__(self, k = None):
super(AssignKBestLabels, self).__init__()
self.k = k
self.copyable_attrs = ['k']
def fit(self, X, y):
"""Fits classifier to training data
Parameters
----------
X : `array_like`, :class:`numpy.matrix` or :mod:`scipy.sparse` matrix, shape=(n_samples, n_features)
input feature matrix
y : `array_like`, :class:`numpy.matrix` or :mod:`scipy.sparse` matrix of `{0, 1}`, shape=(n_samples, n_labels)
binary indicator matrix with label assignments
Returns
-------
self
fitted instance of self
"""
self.n_labels = y.shape[1]
frequencies = (y.sum(axis=0)/float(y.sum().sum())).A.tolist()[0]
labels_sorted_by_frequency = sorted(range(y.shape[1]), key = lambda i: frequencies[i])
self.labels_to_assign = labels_sorted_by_frequency[:self.k]
return self
def predict(self, X):
"""Predict labels for X
Parameters
----------
X : `array_like`, :class:`numpy.matrix` or :mod:`scipy.sparse` matrix, shape=(n_samples, n_features)
input feature matrix
Returns
-------
:mod:`scipy.sparse` matrix of `{0, 1}`, shape=(n_samples, n_labels)
binary indicator matrix with label assignments
"""
prediction = lil_matrix(np.zeros(shape=(X.shape[0], self.n_labels), dtype=int))
prediction[:,self.labels_to_assign] = 1
return prediction
def predict_proba(self, X):
"""Predict probabilities of label assignments for X
Parameters
----------
X : `array_like`, :class:`numpy.matrix` or :mod:`scipy.sparse` matrix, shape=(n_samples, n_features)
input feature matrix
Returns
-------
:mod:`scipy.sparse` matrix of `float in [0.0, 1.0]`, shape=(n_samples, n_labels)
matrix with label assignment probabilities
"""
probabilities = lil_matrix(np.zeros(shape=(X.shape[0], self.n_labels), dtype=float))
probabilities[:,self.labels_to_assign] = 1.0
return probabilities
clf = AssignKBestLabels(k=2)
clf.fit(X_train, y_train)
prediction = clf.predict(X_test)
accuracy_score(y_test, prediction)
Out[99]:
0.10396039603960396
Selecting the base class¶
Madjarov et al. divide approach to multi-label classification into three categories, you should select a scikit-multilearn base class according to the philosophy behind your classifier:
- algorithm adaptation, when a single-label algorithm is directly
adapted to the multi-label case, ex. Decision Trees can be adapted by
taking multiple labels into consideration in decision functions, for
now the base function for this approach is
MLClassifierBase - problem transformation, when the multi-label problem is transformed
to a set of single-label problems, solved there and converted to a
multi-label solution afterwards - for this approach we provide a
comfortable
ProblemTransformationBasebase class - ensemble classification, when the multi-label classification is
performed by an ensemble of multi-label classifiers to improve
performance, overcome overfitting etc. In the case when your
classifier concentrates on clustering the label space, you should use
:class:
LabelSpacePartitioningClassifier- which partitions a label space using a cluster class that implements the :class:LabelSpaceClustererBaseinterface.
Problem transformation approach is centred around the idea of converting a multi-label problem into one or more single-label problems, which are usually solved by single- or multi-class classifiers. Scikit-learn is the de facto standard source of Python implementations of single-label classifiers.
To perform the transformation, every problem transformation classifier needs a base classifier. As all classifiers that follow scikit-s BaseEstimator a clonable, scikit-multilearn’s base class for problem transformation classifiers requires an instance of a base classifier in initialization. Such an instance can be cloned if needed, and its parameters can be set up comfortably.
The biggest problem with joining single-label scikit classifiers with
multi-label classifiers is that there exists no way to learn whether a
given scikit classifier accepts sparse matrices as input for
fit/predict functions. For this reason
ProblemTransformationBase requires another parameter -
require_dense : [ bool, bool ] - a list/tuple of two boolean
values. If the first one is true, that means the base classifier expects
a dense (scikit-compatible array-like of array-likes) representation of
the sample feature space X. If the second one is true - the target
space y is passed to the base classifier as an array like of
numbers. In case any of these are false - the arguments are passed as a
sparse matrix.
If the required_dense argument is not passed, it is set to
[false, false] if a classifier inherits
::class::MLClassifierBase and to [true, true] as a fallback
otherwise. In short, it assumes dense representation is required for
base classifier if the base classifier is not a scikit-multilearn
classifier.
Ensemble classification¶
Ensemble classification is an approach of transforming a multi-label classification problem into a family (an ensemble) of multi-label subproblems.
Unit testing classifiers¶
Scikit-multilearn provides a base unit test class for testing
classifiers. Please check skmultilearn.tests.classifier_basetest for
a general framework for testing the multi-label classifier.
Currently tests test three capabilities of the classifier: - whether the
classifier works with dense/sparse input data
:func:ClassifierBaseTest.assertClassifierWorksWithSparsity - whether
the classifier predicts probabilities using predict_proba for
dense/sparse input data
:func:ClassifierBaseTest.assertClassifierPredictsProbabilities -
whether it is clonable and works with scikit-learn’s cross-validation
classes :func:ClassifierBaseTest.assertClassifierWorksWithCV