6. Multi-label embedding-based classification¶
Multi-label embedding techniques emerged as a response the need to cope with a large label space, but with the rise of computing power they became a method of improving classification quality. Typically the embedding-based multi-label classification starts with embedding the label matrix of the training set in some way, training a regressor for unseen samples to predict their embeddings, and a classifier (sometimes very simple ones) to correct the regression error. Scikit-multilearn provides several multi-label embedders alongisde a general regressor-classifier classification class.
Currently available embedding strategies include:
- Label Network Embeddings via OpenNE network embedding library, as in the LNEMLC paper
- Cost-Sensitive Label Embedding with Multidimensional Scaling, as in the CLEMS paper
- scikit-learn based embeddings such as PCA or manifold learning approaches
Let’s start with loading some data:
In [6]:
import numpy
import sklearn.metrics as metrics
from skmultilearn.dataset import load_dataset
X_train, y_train, feature_names, label_names = load_dataset('emotions', 'train')
X_test, y_test, _, _ = load_dataset('emotions', 'test')
emotions:train - exists, not redownloading
emotions:test - exists, not redownloading
6.1. Label Network Embeddings¶
The label network embeddings approaches require a working tensorflow installation and the OpenNE library. To install them, run the following code:
pip install networkx tensorflow
git clone https://github.com/thunlp/OpenNE/
pip install -e OpenNE/src
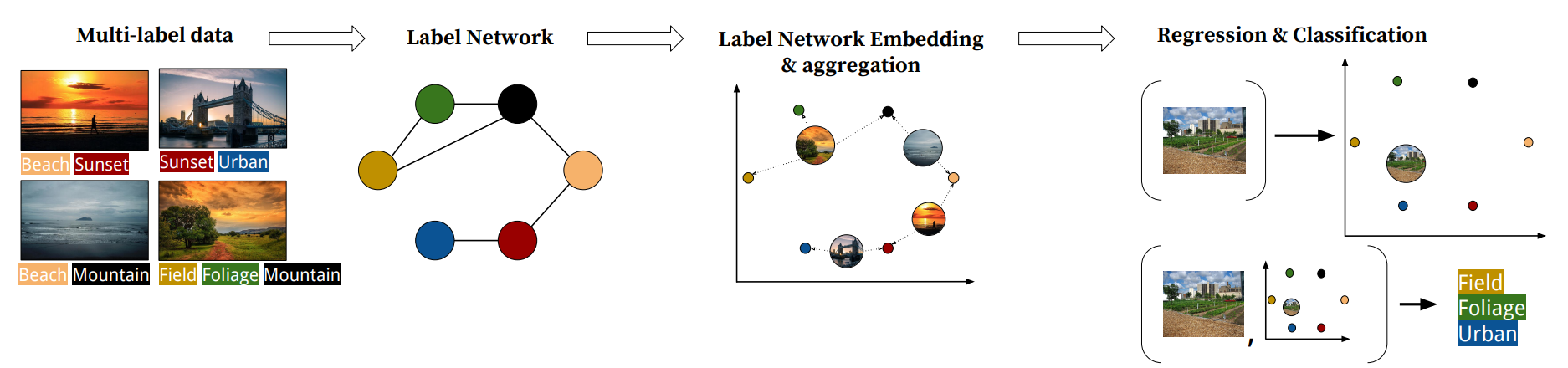
For an example we will use the LINE embedding method, one of the most
efficient and well-performing state of the art approaches, for the
meaning of parameters consult the `OpenNE documentation <>`__. We select
order = 3 which means that the method will take both first and
second order proximities between labels for embedding. We select a
dimension of 5 times the number of labels, as the linear embeddings tend
to need more dimensions for best performance, normalize the label
weights to maintain normalized distances in the network and agregate
label embedings per sample by summation which is a classical approach.
In [7]:
from skmultilearn.embedding import OpenNetworkEmbedder
from skmultilearn.cluster import LabelCooccurrenceGraphBuilder
In [8]:
graph_builder = LabelCooccurrenceGraphBuilder(weighted=True, include_self_edges=False)
openne_line_params = dict(batch_size=1000, order=3)
embedder = OpenNetworkEmbedder(
graph_builder,
'LINE',
dimension = 5*y_train.shape[1],
aggregation_function = 'add',
normalize_weights=True,
param_dict = openne_line_params
)
We now need to select a regressor and a classifier, we use random forest regressors with MLkNN which is a well working combination often used for multi-label embedding:
In [9]:
from skmultilearn.embedding import EmbeddingClassifier
from sklearn.ensemble import RandomForestRegressor
from skmultilearn.adapt import MLkNN
In [10]:
clf = EmbeddingClassifier(
embedder,
RandomForestRegressor(n_estimators=10),
MLkNN(k=5)
)
clf.fit(X_train, y_train)
predictions = clf.predict(X_test)
Pre-procesing for non-uniform negative sampling!
Pre-procesing for non-uniform negative sampling!
epoch:0 sum of loss:4.97153359652
epoch:0 sum of loss:5.11869335175
epoch:1 sum of loss:4.98133981228
epoch:1 sum of loss:4.97720247507
epoch:2 sum of loss:4.81723511219
epoch:2 sum of loss:5.05428689718
epoch:3 sum of loss:5.09079384804
epoch:3 sum of loss:4.72988516092
epoch:4 sum of loss:5.0347994566
epoch:4 sum of loss:4.95063251257
epoch:5 sum of loss:4.68008613586
epoch:5 sum of loss:4.9329983592
epoch:6 sum of loss:4.74205821753
epoch:6 sum of loss:4.68989795446
epoch:7 sum of loss:4.62912601233
epoch:7 sum of loss:4.81548637152
epoch:8 sum of loss:4.40033769608
epoch:8 sum of loss:4.73801320791
epoch:9 sum of loss:4.61178982258
epoch:9 sum of loss:4.91443294287
6.2. Cost-Sensitive Label Embedding with Multidimensional Scaling¶
CLEMS is another well-perfoming method in multi-label embeddings. It
uses weighted multi-dimensional scaling to embedd a cost-matrix of
unique label combinations. The cost-matrix contains the cost of
mistaking a given label combination for another, thus real-valued
functions are better ideas than discrete ones. Also, the is_score
parameter is used to tell the embedder if the cost function is a score
(the higher the better) or a loss (the lower the better). Additional
params can be also assigned to the weighted scaler. The most efficient
parameter for the number of dimensions is equal to number of labels, and
is thus enforced here.
In [14]:
from skmultilearn.embedding import CLEMS, EmbeddingClassifier
from sklearn.ensemble import RandomForestRegressor
from skmultilearn.adapt import MLkNN
dimensional_scaler_params = {'n_jobs': -1}
clf = EmbeddingClassifier(
CLEMS(metrics.jaccard_similarity_score, is_score=True, params=dimensional_scaler_params),
RandomForestRegressor(n_estimators=10, n_jobs=-1),
MLkNN(k=1),
regressor_per_dimension= True
)
clf.fit(X_train, y_train)
predictions = clf.predict(X_test)
6.3. Scikit-learn based embedders¶
Any scikit-learn embedder can be used for multi-label classification embeddings with scikit-multilearn, just select one and try, here’s a spectral embedding approach with 10 dimensions of the embedding space:
In [15]:
from skmultilearn.embedding import SKLearnEmbedder, EmbeddingClassifier
from sklearn.manifold import SpectralEmbedding
from sklearn.ensemble import RandomForestRegressor
from skmultilearn.adapt import MLkNN
clf = EmbeddingClassifier(
SKLearnEmbedder(SpectralEmbedding(n_components = 10)),
RandomForestRegressor(n_estimators=10),
MLkNN(k=5)
)
clf.fit(X_train, y_train)
predictions = clf.predict(X_test)